GCN Deep Graph Infomax on CORA¶
This demo demonstrates how to perform unsupervised training of a GCN, GAT, APPNP, or GraphSAGE model using the Deep Graph Infomax algorithm (https://arxiv.org/pdf/1809.10341.pdf) on the CORA dataset.
As with all StellarGraph workflows: first we load the dataset, next we create our data generators, and then we train our model. We then take the embeddings created through unsupervised training and predict the node classes using logistic regression.
Run the master version of this notebook: |
[1]:
# install StellarGraph if running on Google Colab
import sys
if 'google.colab' in sys.modules:
%pip install -q stellargraph[demos]==1.0.0rc1
[2]:
# verify that we're using the correct version of StellarGraph for this notebook
import stellargraph as sg
try:
sg.utils.validate_notebook_version("1.0.0rc1")
except AttributeError:
raise ValueError(
f"This notebook requires StellarGraph version 1.0.0rc1, but a different version {sg.__version__} is installed. Please see <https://github.com/stellargraph/stellargraph/issues/1172>."
) from None
[3]:
from stellargraph.mapper import (
CorruptedGenerator,
FullBatchNodeGenerator,
GraphSAGENodeGenerator,
HinSAGENodeGenerator,
)
from stellargraph import StellarGraph
from stellargraph.layer import GCN, DeepGraphInfomax, GraphSAGE, GAT, APPNP, HinSAGE
from stellargraph import datasets
from stellargraph.utils import plot_history
import pandas as pd
from matplotlib import pyplot as plt
from sklearn import model_selection
from sklearn.linear_model import LogisticRegression
from sklearn.manifold import TSNE
from IPython.display import display, HTML
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import EarlyStopping
import tensorflow as tf
from tensorflow.keras import Model
[4]:
dataset = datasets.Cora()
display(HTML(dataset.description))
G, node_subjects = dataset.load()
## Data Generators
Now we create the data generators using CorruptedGenerator. CorruptedGenerator returns shuffled node features along with the regular node features and we train our model to discriminate between the two.
Note that:
- We typically pass all nodes to
corrupted_generator.flowbecause this is an unsupervised task - We don’t pass
targetstocorrupted_generator.flowbecause these are binary labels (true nodes, false nodes) that are created byCorruptedGenerator
[5]:
fullbatch_generator = FullBatchNodeGenerator(G, sparse=False)
gcn_model = GCN(layer_sizes=[128], activations=["relu"], generator=fullbatch_generator)
corrupted_generator = CorruptedGenerator(fullbatch_generator)
gen = corrupted_generator.flow(G.nodes())
Using GCN (local pooling) filters...
Model Creation and Training¶
We create and train our DeepGraphInfomax model. Note that the loss used here must always be tf.nn.sigmoid_cross_entropy_with_logits.
[6]:
infomax = DeepGraphInfomax(gcn_model, corrupted_generator)
x_in, x_out = infomax.in_out_tensors()
model = Model(inputs=x_in, outputs=x_out)
model.compile(loss=tf.nn.sigmoid_cross_entropy_with_logits, optimizer=Adam(lr=1e-3))
[7]:
epochs = 100
[8]:
es = EarlyStopping(monitor="loss", min_delta=0, patience=20)
history = model.fit(gen, epochs=epochs, verbose=0, callbacks=[es])
plot_history(history)
['...']
Extracting Embeddings and Logistic Regression¶
Since we’ve already trained the weights of our base model - GCN in this example - we can simply use base_model.in_out_tensors to obtain the trained node embedding model. Then we use logistic regression on the node embeddings to predict which class the node belongs to.
Note that the results here differ from the paper due to different train/test/val splits.
[9]:
x_emb_in, x_emb_out = gcn_model.in_out_tensors()
# for full batch models, squeeze out the batch dim (which is 1)
x_out = tf.squeeze(x_emb_out, axis=0)
emb_model = Model(inputs=x_emb_in, outputs=x_out)
[10]:
train_subjects, test_subjects = model_selection.train_test_split(
node_subjects, train_size=0.1, test_size=None, stratify=node_subjects
)
test_gen = fullbatch_generator.flow(test_subjects.index)
train_gen = fullbatch_generator.flow(train_subjects.index)
test_embeddings = emb_model.predict(test_gen)
train_embeddings = emb_model.predict(train_gen)
lr = LogisticRegression(multi_class="auto", solver="lbfgs")
lr.fit(train_embeddings, train_subjects)
y_pred = lr.predict(test_embeddings)
gcn_acc = (y_pred == test_subjects).mean()
print(f"Test classification accuracy: {gcn_acc}")
Test classification accuracy: 0.7981952420016407
This accuracy is close to that for training a supervised GCN model end-to-end, suggesting that Deep Graph Infomax is an effective method for unsupervised training.
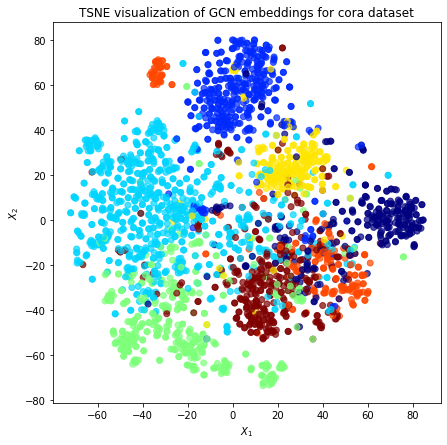
Visualisation with TSNE¶
Here we visualize the node embeddings with TSNE. As you can see below, the Deep Graph Infomax model produces well separated embeddings using unsupervised training.
[11]:
all_embeddings = emb_model.predict(fullbatch_generator.flow(G.nodes()))
y = node_subjects.astype("category")
trans = TSNE(n_components=2)
emb_transformed = pd.DataFrame(trans.fit_transform(all_embeddings), index=G.nodes())
emb_transformed["label"] = y
[12]:
alpha = 0.7
fig, ax = plt.subplots(figsize=(7, 7))
ax.scatter(
emb_transformed[0],
emb_transformed[1],
c=emb_transformed["label"].cat.codes,
cmap="jet",
alpha=alpha,
)
ax.set(aspect="equal", xlabel="$X_1$", ylabel="$X_2$")
plt.title("TSNE visualization of GCN embeddings for cora dataset")
plt.show()
Comparing Different Models¶
Now we run Deep Graph Infomax training for GAT, GCN, APPNP, and GraphSAGE. Note that switching between StellarGraph models only requires a few code changes.
[13]:
def run_deep_graph_infomax(base_model, generator, epochs):
corrupted_generator = CorruptedGenerator(generator)
gen = corrupted_generator.flow(G.nodes())
infomax = DeepGraphInfomax(base_model, corrupted_generator)
x_in, x_out = infomax.in_out_tensors()
model = Model(inputs=x_in, outputs=x_out)
model.compile(loss=tf.nn.sigmoid_cross_entropy_with_logits, optimizer=Adam(lr=1e-3))
history = model.fit(gen, epochs=epochs, verbose=0, callbacks=[es])
x_emb_in, x_emb_out = base_model.in_out_tensors()
# for full batch models, squeeze out the batch dim (which is 1)
if isinstance(base_model, (GAT, GCN, APPNP)):
x_emb_out = tf.squeeze(x_emb_out, axis=0)
emb_model = Model(inputs=x_emb_in, outputs=x_emb_out)
test_gen = generator.flow(test_subjects.index)
train_gen = generator.flow(train_subjects.index)
test_embeddings = emb_model.predict(test_gen)
train_embeddings = emb_model.predict(train_gen)
lr = LogisticRegression(multi_class="auto", solver="lbfgs")
lr.fit(train_embeddings, train_subjects)
y_pred = lr.predict(test_embeddings)
acc = (y_pred == test_subjects).mean()
return acc
[14]:
gat_model = GAT(
layer_sizes=[128], activations=["relu"], generator=fullbatch_generator, attn_heads=8,
)
gat_acc = run_deep_graph_infomax(gat_model, fullbatch_generator, epochs=epochs)
gat_acc
print(f"Test classification accuracy: {gat_acc}")
['...']
Test classification accuracy: 0.448318293683347
[15]:
appnp_model = APPNP(
layer_sizes=[128], activations=["relu"], generator=fullbatch_generator
)
appnp_acc = run_deep_graph_infomax(appnp_model, fullbatch_generator, epochs=epochs)
print(f"Test classification accuracy: {appnp_acc}")
['...']
Test classification accuracy: 0.4470877768662838
[16]:
graphsage_generator = GraphSAGENodeGenerator(G, batch_size=1000, num_samples=[5])
graphsage_model = GraphSAGE(
layer_sizes=[128], activations=["relu"], generator=graphsage_generator
)
graphsage_acc = run_deep_graph_infomax(
graphsage_model, graphsage_generator, epochs=epochs
)
print(f"Test classification accuracy: {graphsage_acc}")
['...']
Test classification accuracy: 0.7013945857260049
Cora is a homogeneous graph, with only one type of node (paper) and one type of edge (type). Models designed for heterogeneous graphs (with moer than one of either) can also be applied to homogeneous graphs, but it is not using their additional flexibility.
HinSAGE is a generalisation of GraphSAGE to heterogeneous graphs that can be trained with Deep Graph Infomax. For homogeneous graphs, it is equivalent to GraphSAGE and it indeed gives similar results.
[17]:
hinsage_generator = HinSAGENodeGenerator(
G, batch_size=1000, num_samples=[5], head_node_type="paper"
)
hinsage_model = HinSAGE(
layer_sizes=[128], activations=["relu"], generator=hinsage_generator
)
hinsage_acc = run_deep_graph_infomax(hinsage_model, hinsage_generator, epochs=epochs)
print(f"Test classification accuracy: {hinsage_acc}")
['...']
Test classification accuracy: 0.7038556193601313
The cell below shows the accuracy of each model.
[18]:
pd.DataFrame(
[gat_acc, gcn_acc, appnp_acc, graphsage_acc, hinsage_acc],
index=["GAT", "GCN", "APPNP", "GraphSAGE", "HinSAGE"],
columns=["Accuracy"],
)
[18]:
| Accuracy | |
|---|---|
| GAT | 0.448318 |
| GCN | 0.798195 |
| APPNP | 0.447088 |
| GraphSAGE | 0.701395 |
| HinSAGE | 0.703856 |
Run the master version of this notebook: |